Product specifications
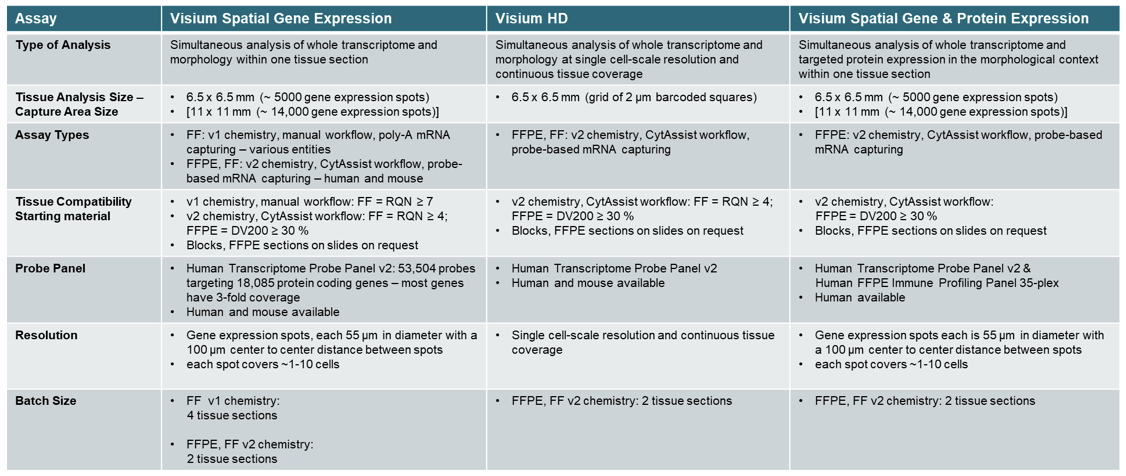
Sequencing based whole transcriptome analysis in the morphological context, targeting 18,000 human and 20,000 mouse genes (Visium CytAssist probe-based assays)
Compatibility:
- Visium CytAssist probe-based assays for human and mouse Fresh Frozen or FFPE blocks, pre-sectioned FFPE tissue on glass slides
- Visium PolyA capture-based direct placement method for Fresh Frozen (variable species) Tissue Analysis Size & Resolution:
- Standard Visium Slides with capture areas of 6.5 x 6.5 mm (~ 5000 gene expression spots) or 11 x 11 mm (~ 14,000 gene expression spots), barcoded gene expression spots are 55 µm in diameter and 100 µm distance from center to center between spots, providing an average resolution of 1 to 10 cells per spot.
- Visium HD Slides with capture areas of 6.5 x 6.5 mm with a continuous lawn of oligonucleotides in a grid of 2 µm barcoded squares, no gaps between barcoded oligonucleotids providing a single-scale resolution
- Standard Visium Slides with capture areas of 6.5 x 6.5 mm (~ 5000 gene expression spots) or 11 x 11 mm (~ 14,000 gene expression spots), barcoded gene expression spots are 55 µm in diameter and 100 µm distance from center to center between spots, providing an average resolution of 1 to 10 cells per spot.

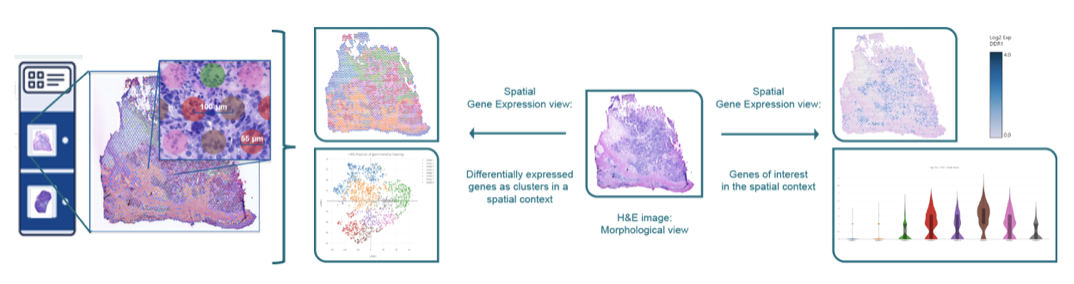
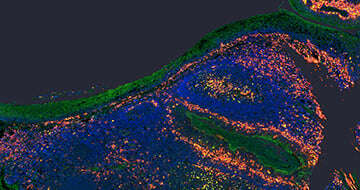
 10x Genomics Visium technology offers groundbreaking spatial gene expression profiling. This cutting-edge platform enables researchers to visualize and analyze gene expression directly within the tissue’s spatial context. By integrating spatial and molecular data, our services empower you to uncover critical biological insights that are impossible with traditional bulk or single-cell RNA sequencing methods. By combining histology and whole transcriptome profiling (using fresh frozen or FFPE), this technology allows you to spatially map gene expression within tissues, providing valuable insights into cellular function and interactions. Whether you’re investigating tumor microenvironments, tumor heterogeneity, developmental biology, or other complex systems, our solution provides the precision and clarity needed to advance your discoveries.
10x Genomics Visium technology offers groundbreaking spatial gene expression profiling. This cutting-edge platform enables researchers to visualize and analyze gene expression directly within the tissue’s spatial context. By integrating spatial and molecular data, our services empower you to uncover critical biological insights that are impossible with traditional bulk or single-cell RNA sequencing methods. By combining histology and whole transcriptome profiling (using fresh frozen or FFPE), this technology allows you to spatially map gene expression within tissues, providing valuable insights into cellular function and interactions. Whether you’re investigating tumor microenvironments, tumor heterogeneity, developmental biology, or other complex systems, our solution provides the precision and clarity needed to advance your discoveries.